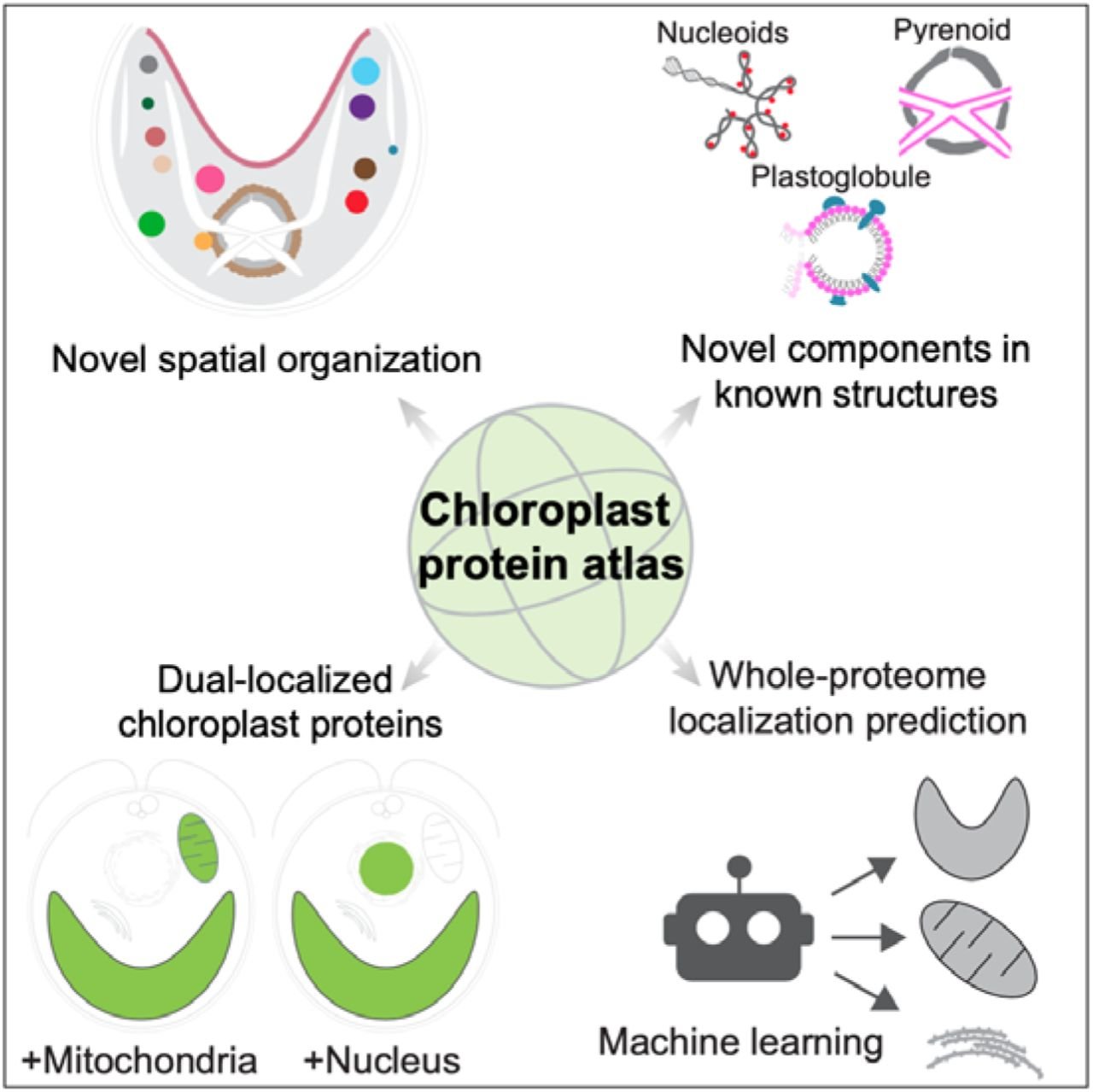
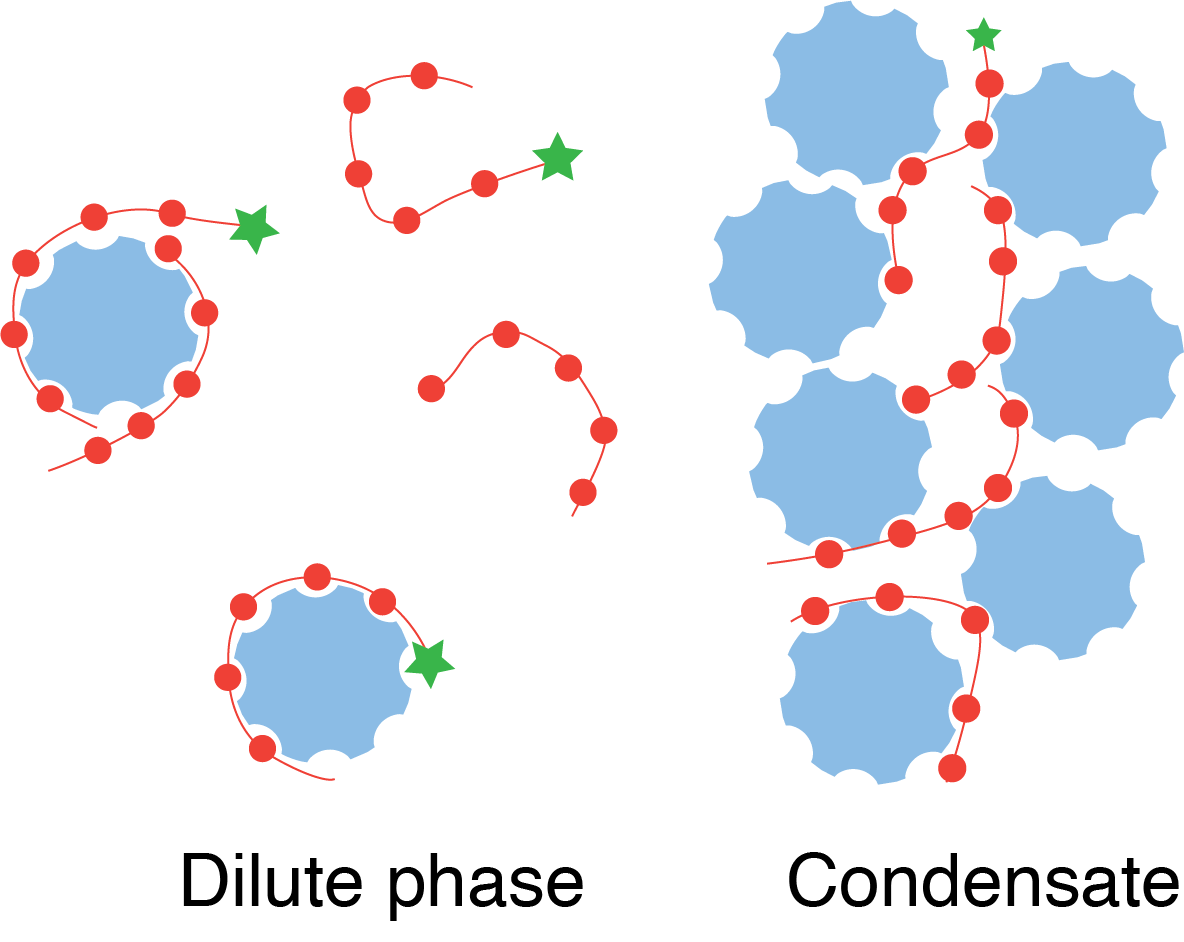
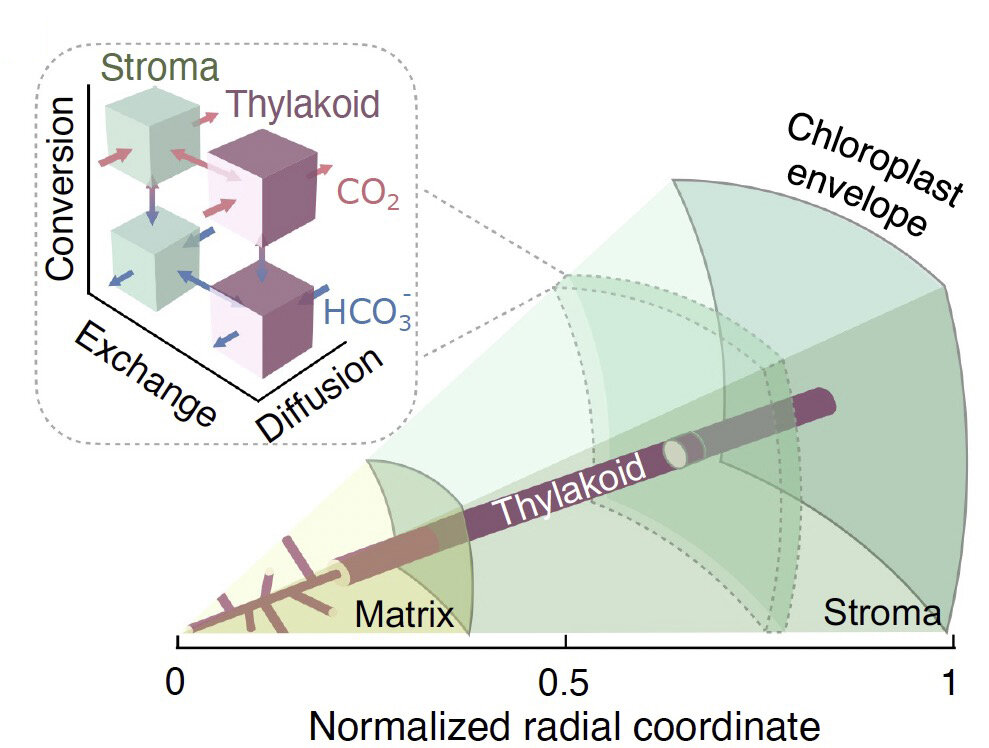
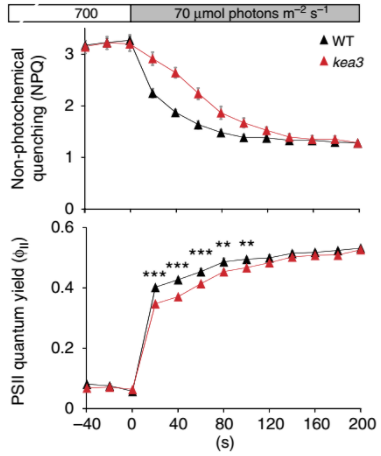
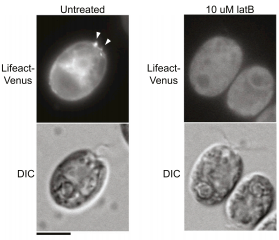
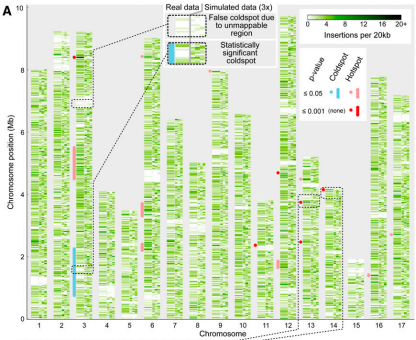
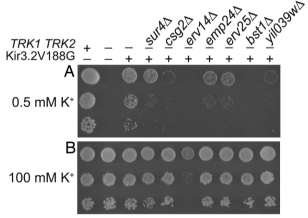
Jonikas Lab member names are bolded.
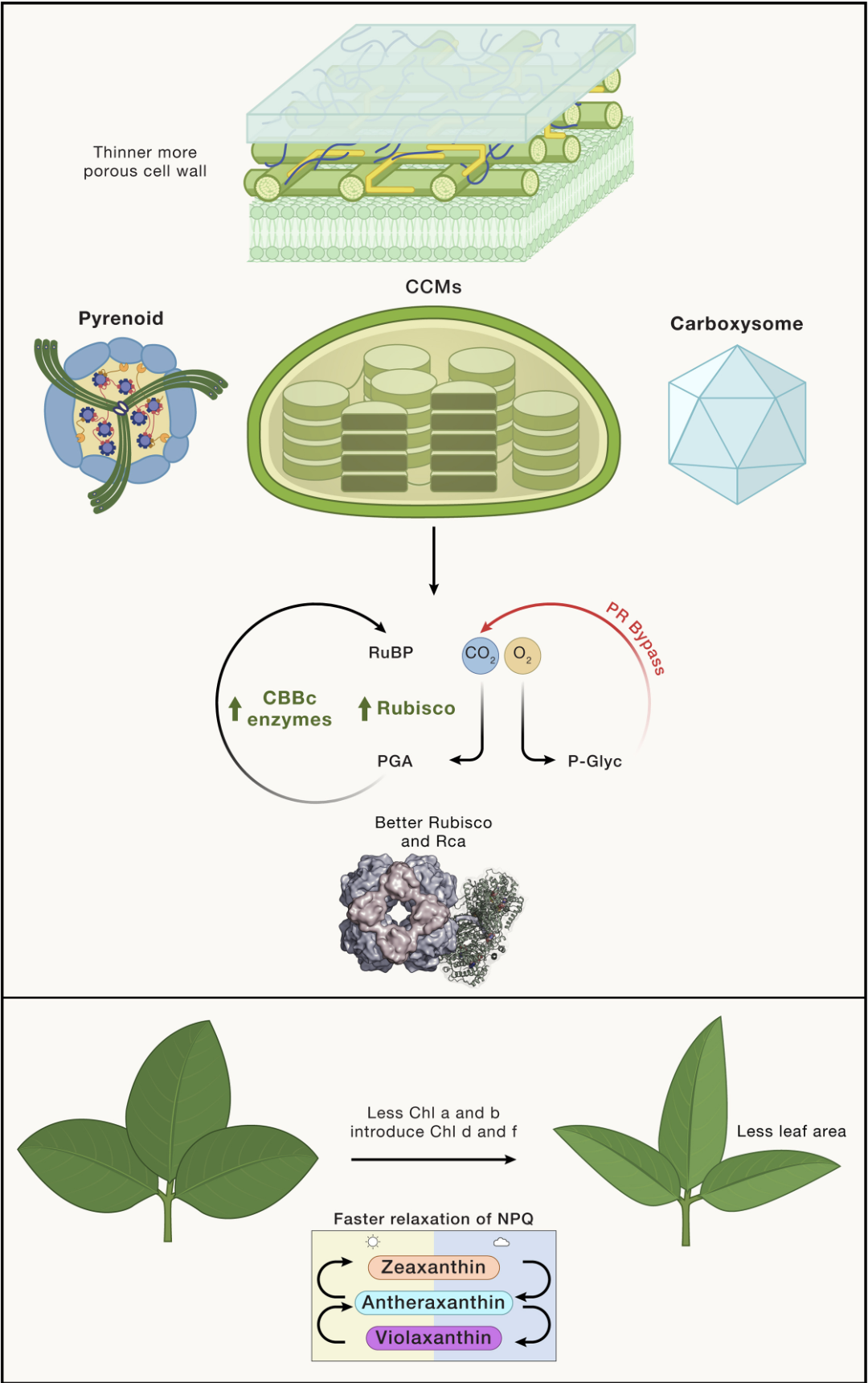
Feeding from the sun—Successes and prospects in bioengineering photosynthesis for food security
Long SP, Wang Y, Carmo-Silva E, Cavanagh AP, Jonikas MC, Kromdijk J, Long BM, Marshall-Colón A, Shukla D, Wilson RH, Zhu X, & Ainsworth EA
Cell (2025) 188:6700 - 6719 | https://doi.org/10.1016/j.cell.2025.10.033
The Arabidopsis amino acid transporter UmamiT20 confers Botrytis cinerea susceptibility
Prior MJ, Weidauer D, Locci F, Liao J, Kuwata K, Deng C, Ye HB, Cai Q, Bezrutczyk M, Zhao C, Jonikas MC, Pilot G, Jin H, Parker JE, Frommer WB, & Kim J
Journal of Experimental Botany (2025) accepted manuscript | https://doi.org/10.1093/jxb/eraf496
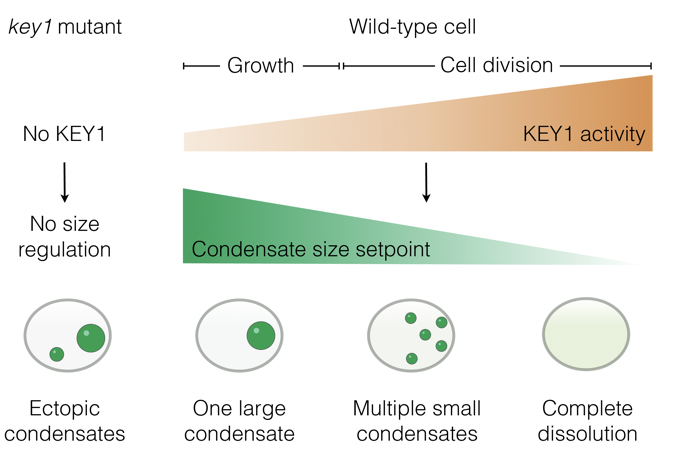
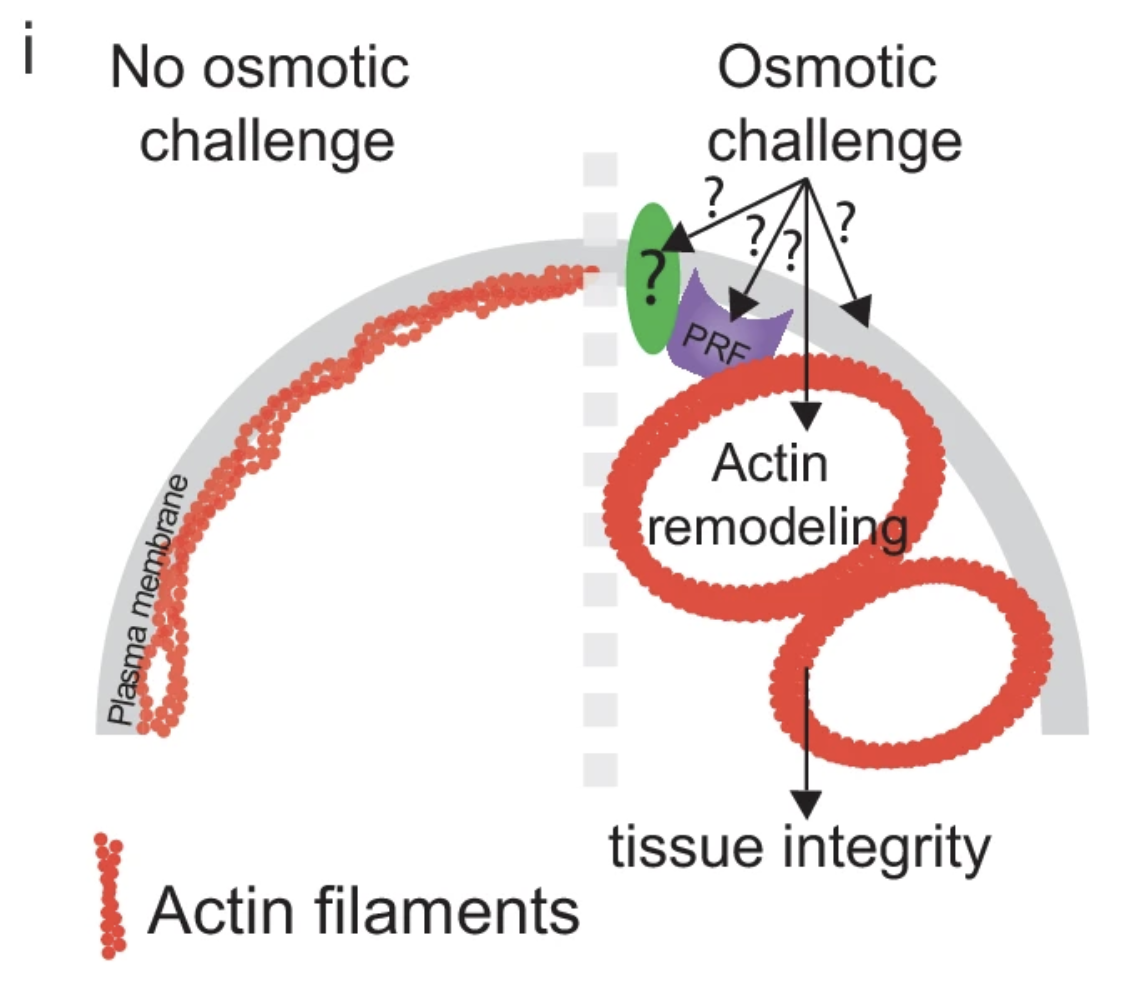
*He S, *Lemma LM, Martinez-Calvo A, He G, Hennacy JH, Wang L, Ergun SL, Rai AK, Wang C, Bunday L, Kayser-Browne A, Wang Q, Brangwynne CP, #Wingreen NS, & #Jonikas MC
*Equal Contribution, # Corresponding Authors
Preprint (2025) bioRxiv | https://doi.org/10.1101/2025.10.09.681382
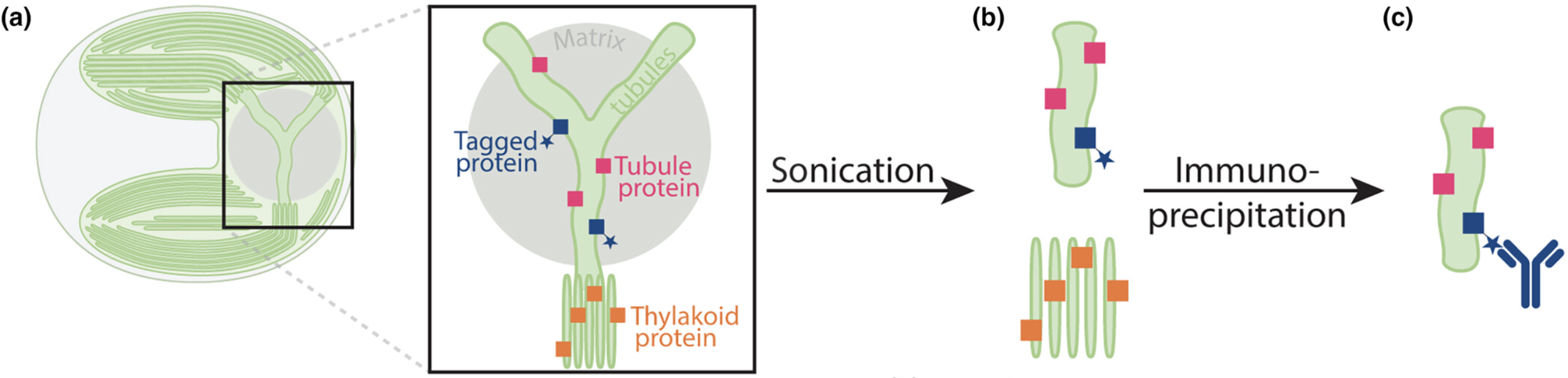
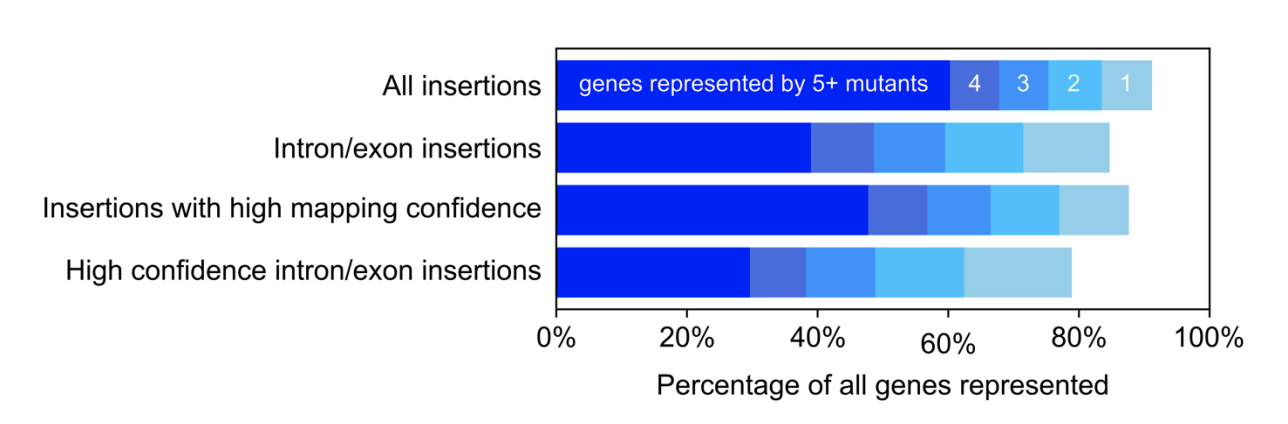
Franklin E, Wang L, Cruz ER, Duggal K, Ergun SL, Garde A, Lunardon A, Patena W, Pacini C, & Jonikas MC
New Phytologist (2025) 4:249 | https://doi.org/10.1111/nph.70669
PMID: 41116735
Fei C, Wingreen NS, & Jonikas MC
Plant Physiology (2025) 4:198 | https://doi.org/10.1093/plphys/kiaf316
PMID: 40794800
Lunardon A*, Patena W*, Pacini C, Warren-Williams M, Zubak Y, Laudon M, Silflow C, Lefebvre P, Jonikas MC
*Equal Contribution
Preprint (2024) | https://doi.org/10.1101/2024.12.16.626622
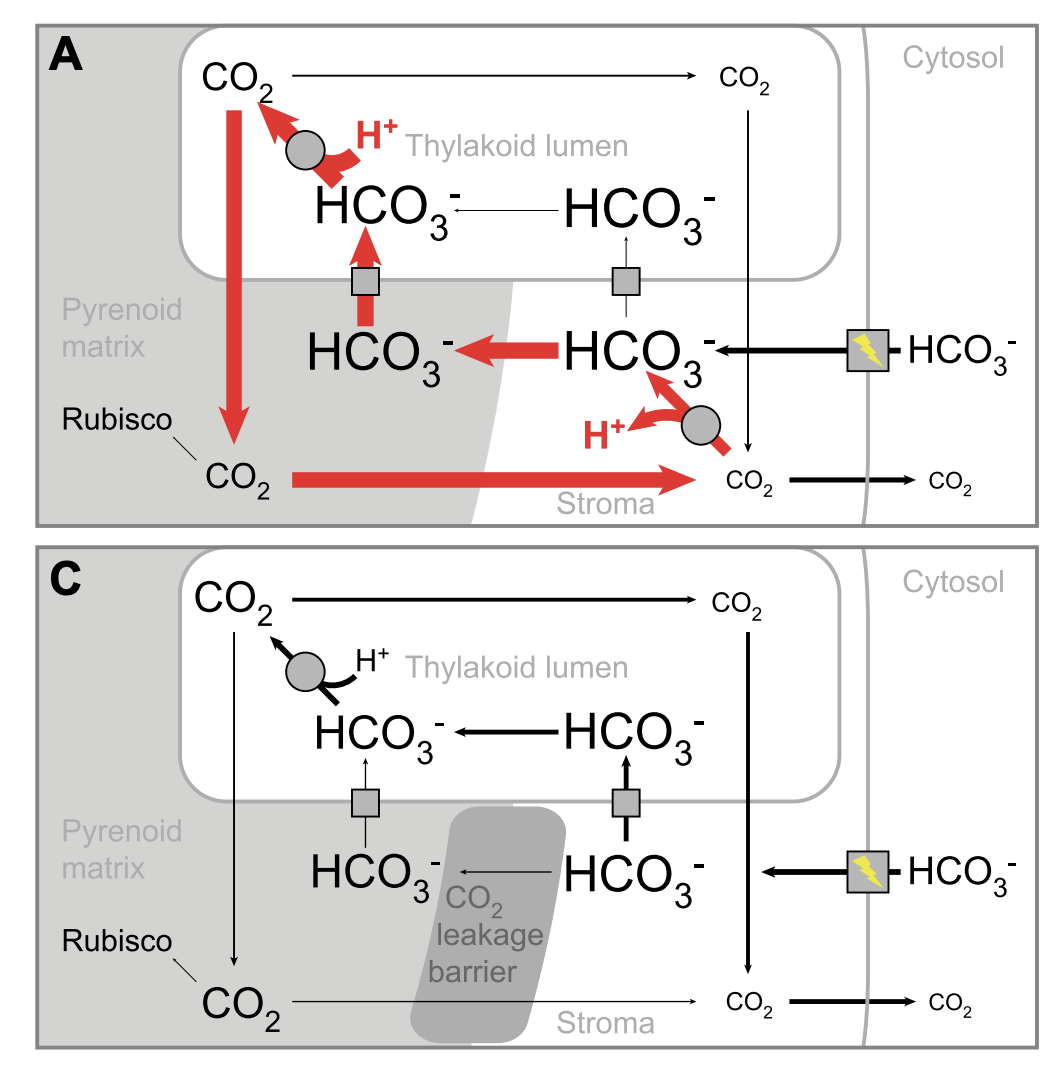
SAGA1 and MITH1 produce matrix-traversing membranes in the CO2-fixing pyrenoid
Hennacy JH, Atkinson N, Kayser-Browne A, Ergun SL, Franklin E, Wang L, Eicke S, Kazachkova Y, Kafri M, Fauser F, Vilarrasa-Blasi J, Jinkerson RE, Zeeman SC, McCormick AJ & Jonikas MC
Nature Plants (2024) 10:2038–2051 | https://doi: 10.1038/s41477-024-01847-0
PMID: 39548241
Vilarrasa-Blasi J, Vellosillo T, Jinkerson RE, Fauser F, Xiang T, Minkoff BB, Wang L, Kniazev K, Guzman M, Osaki J, Barrett-Wilt GA, Sussman M, Jonikas MC, & Dinneny J
Nature Communications (2024) 15:1 | https://doi.org/10.1038/s41467-024-49844-3
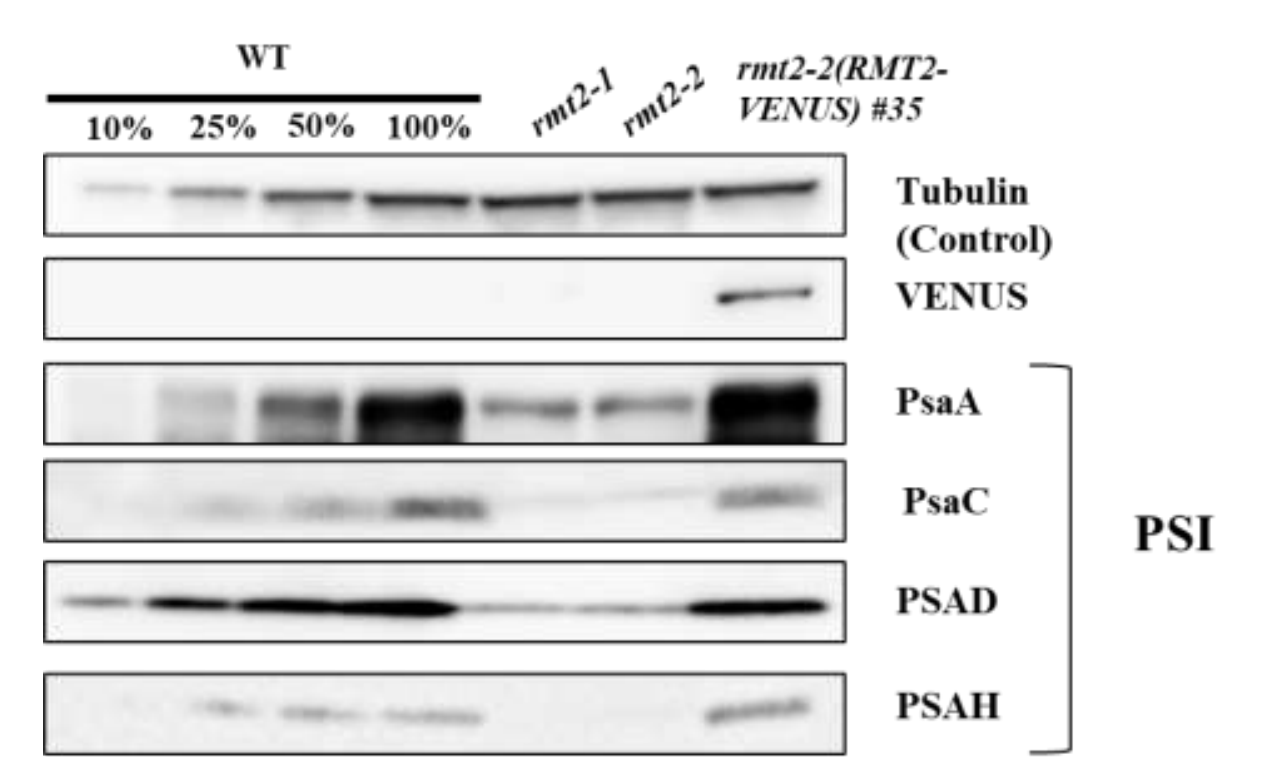
Chloroplast Methyltransferase Homolog RMT2 is Involved in Photosystem I Biogenesis
Kima RG, Huanga W, Findiniera J, Bunburya F, Redekopa P, Shresthaa R, Grismera TS, Vilarrasa-Blasib J, Jinkersonc RE, Fakhimia N, Fausera F, Jonikas MC, Onishie M, Xua S, and Grossman AR
Preprint bioRxiv | https://doi.org/10.1101/2023.12.21.572672
Tóth D, Kuntam S, Ferenczi Á, Vidal-Meireles A, Kovács L, Wang L, Sarkadi Z, Migh E, Szentmihályi K, Tengölics R, Neupert J, Bock R, Jonikas MC, Molnar A, & Tóth SZ
Plant Physiology (2023) 194: 1646–1661 | https://doi.org/10.1093/plphys/kiad607
PMID: 37962583
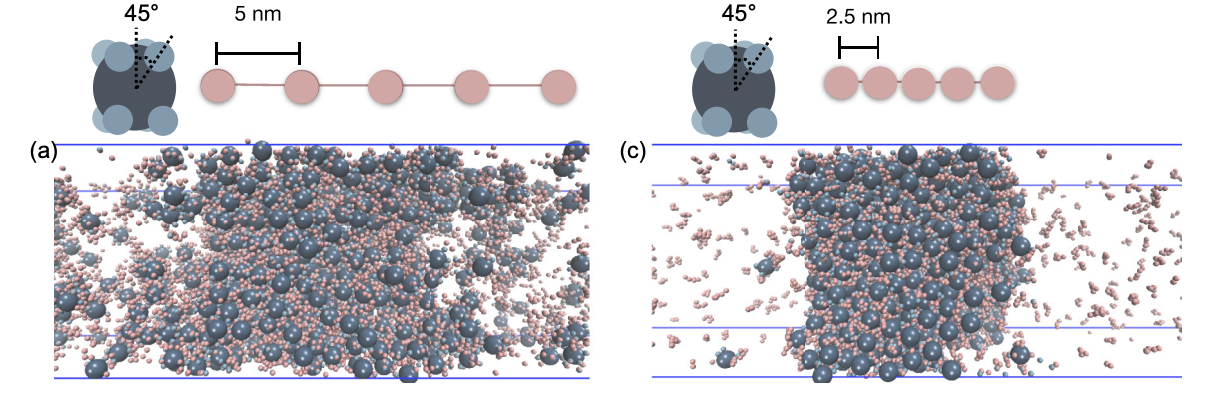
Impact of Linker Length on Biomolecular Condensate Formation
GrandPre T, Zhang Y, Pyo AGT, Weiner B, Li J, Jonikas MC, & Wingreen NS
PRX Life (2023) 1:023013 | https://doi.org/10.1103/PRXLife.1.023013
Kafri M, Patena W, Martin L, Wang L, Gomer G, Ergun SL, Sirkejyan AK, Goh A, Wilson AT, Gavrilenko SE, Breker M, Roichman A, McWhite CD, Rabinowitz JD, Cross FR, Wühr M, & Jonikas MC.
Cell (2023) 186: 5638-5655 | https://doi: 10.1016/j.cell.2023.11.007
PMID: 38065083
The pyrenoid: the eukaryotic CO2-concentrating organelle
He S, Crans VL, Jonikas MC.
The Plant Cell (2023) 35:3236–3259 | https://doi.org/10.1093/plcell/koad157
PMID: 37279536
Wang L, Patena W, Van Baalen KA, Xie Y, Singer ER, Gavrilenko S, Warren-Williams M, Han L, Harrigan HR, Hartz LD, Chen V, Ton VTNP, Kyin S, Shwe HH, Cahn MH, Wilson AT, Hu J, Schnell DJ, McWhite CD, & Jonikas MC.
Cell (2023) 186:3499–3518 | https://doi.org/10.1016/j.cell.2023.06.008
PMID: 37437571
Phase-separating pyrenoid proteins form complexes in the dilute phase
He G, GrandPre T, Wilson H, Zhang Y, #Jonikas MC, #Wingreen NS, & #Wang Q.
#Corresponding authors
Communications Biology (2023) 6: 19 | https://doi.org/10.1038/s42003-022-04373-x
PMID: 36611062
*Fei C, *Wilson AT, #Mangan NM, #Wingreen NS, & #Jonikas MC.
*Equal contribution #Corresponding authors
Nature Plants (2022) 8:583–595
PMID: 35596080
Systematic characterization of gene function in the photosynthetic alga Chlamydomonas reinhardtii
*Fauser F, *Vilarrasa-Blasi J, Onishi M, Ramundo S, Patena W, Millican M, Osaki J, Philp C, Nemeth M, Salomé PA, Li X, Wakao S, Kim RG, Kaye Y, Grossman AR, Niyogi KK, Merchant S, Cutler S, Walter P, #Dinneny JR, #Jonikas MC, & #Jinkerson RE.
*Equal contribution #Corresponding authors
Nature Genetics (2022) 54:705–714
PMID: 35513725
Ramundo S, Asakura Y, Salomé PA, Strenkert D, Boone M, Mackinder LCM, Takafuji K, Dinc E, Rahire M, Crèvecoeur M, Magneschi L, Schaad O, Hippler M, Jonikas MC, Merchant S, #Nakai M, #Rochaix JD, & #Walter P.
#Corresponding authors
Proceedings of the National Academy of Sciences U S A (2020) 3:202014294. doi: 10.1073/pnas.2014294117.
PMID: 33273113
Increasing the uptake of carbon dioxide
Franklin E & Jonikas M.
eLife (2020) 9:e64380. doi: 10.7554/eLife.64380.
PMID: 33270556
The structural basis of Rubisco phase separation in the pyrenoid
He S, Chou HT, Matthies D, Wunder T, Meyer MT, Atkinson N, Martinez-Sanchez A, Jeffrey PD, Port SA, Patena W, He G, Chen VK, Hughson FM, McCormick AJ, Mueller-Cajar O, Engel BD, Yu Z, & Jonikas MC.
Nature Plants (2020) 6:1480-1490.
PMID: 33230314
Assembly of the algal CO2-fixing organelle, the pyrenoid, is guided by a Rubisco-binding motif
Meyer MT, Itakura AK, Patena W, Wang L, He S, Emrich-Mills T, Lau CS, Yates G, Mackinder LCM, & Jonikas MC.
Science Advances (2020) 6:eabd2408
PMID: 33177094
Rigidity enhances a magic-number effect in polymer phase separation
Xu B, He G, Weiner BG, Ronceray P, Meir Y, Jonikas MC, & Wingreen NS.
Nature Communications (2020) 11:1561.
PMID: 32214099
Hennacy JH & Jonikas MC.
Annual Review of Plant Biology (2020) 71:461-485
PMID: 32151155
*Itakura AK, *Chan KX, Atkinson N, Pallesen L, Wang L, Reeves G, Patena W, Caspari O, Roth R, Goodenough U, McCormick AJ, #Griffiths H, & #Jonikas MC.
*Equal contribution #Corresponding authors
Proceedings of the National Academy of Sciences (2019) 116: 18445-18454.
PMID: 31455733
Perlaza K, Toutkoushian H, Boone M, Lam M, Iwai M, Jonikas MC, #Walter P, & #Ramundo S.
#Corresponding authors
eLife (2019) doi: 10.7554/eLife.49577.001
PMID: 31612858
Li X, Patena W, Fauser F, Jinkerson RE, Saroussi S, Meyer MT, Ivanova N, Robertson JM, Yue R, Zhang R, Vilarrasa-Blasi J, Wittkopp TM, Ramundo S, Blum SR, Goh A, Laudon M, Srikumar T, Lefebvre PA, Grossman AR, & Jonikas MC.
Nature Genetics (2019) 51:627-635
PMID: 30886426
Phase separation in biology and disease-a symposium report
Cable J, Brangwynne C, Seydoux G, Cowburn D, Pappu RV, Castañeda CA, Berchowitz LE, Chen Z, Jonikas M, Dernburg A, Mittag T, Fawzi NL.
Annals of the New York Academy of Sciences (2019) 1452: 3-11.
PMID: 31199001
Küken A, Sommer F, Yaneva-Roder L, Mackinder LC, Höhne M, Geimer S, Jonikas MC, Schroda M, Stitt M, Nikoloski Z, & Mettler-Altmann T.
eLife (2018) doi: 10.7554/eLife.37960.
PMID: 30306890
A Spatial Interactome Reveals the Protein Organization of the Algal CO2-Concentrating Mechanism
Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, & Jonikas MC.
Cell (2017) 171: 133-147.
PMID: 28938113
Highlighted on the cover of Cell and in a Cell Preview by Jean-David Rochaix.
The Eukaryotic CO2-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization
Freeman ES, Xu B†, Kuhn Cuellar L†, Martinez-Sanchez A, Schaffer M, Strauss M, Cartwright HN, Ronceray P, Plitzko JM, Förster F, #Wingreen NS, #Engel BD, ‡Mackinder LCM, & ‡#Jonikas MC.
†Equal contribution ‡Equal contribution #Corresponding authors
Cell (2017) 171: 148-162.
PMID: 28938114
Highlighted in a Research Highlight in Nature Plants.
A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle
Mackinder LCM, Meyer MT, Mettler-Altmann T, Chen VK, Mitchell MC, Caspari O, Freeman Rosensweig ES, Pallesen L, Reeves G, Itakura A, Roth R, Sommer F, Geimer S, Mühlhaus T, Schroda M, Goodenough U, Stitt M, Griffiths H, & Jonikas MC.
Proceedings of the National Academy of Sciences U S A (2016) 113: 5958-63.
PMID: 27166422
Armbruster U, Leonelli L, Correa Galvis V, Strand D, Quinn EH, Jonikas MC, & Niyogi KK.
Plant and Cell Physiology (2016) 57: 1557-1567
PMID: 27335350
Li X & Jonikas MC.
Chapter 10 in Lipids in Plant and Algae Development, Y. Nakamura, Y. Li-Beisson (eds.), (2016) Springer.
PMID: 27023238
An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii.
*Li X, *Zhang R, *Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre P, Fitz-Gibbon ST, Grossman AR, & Jonikas MC.
*Equal contribution
The Plant Cell. (2016) 28: 367-87.
PMID: 26764374
*Yang W, *Wittkopp TM, Li X, Warakanont J, Dubini A, Catalanotti C, Kim RG, Nowack EC, Mackinder LC, Aksoy M, Page MD, D'Adamo S, Saroussi S, Heinnickel M, Johnson X, Richaud P, Alric J, Boehm M, Jonikas MC, Benning C, Merchant SS, Posewitz MC, & Grossman AR.
*Equal contribution
Proceedings of the National Academy of Sciences U S A (2015) 112:14978-83.
PMID: 26627249
Atkinson N, Feike D, Mackinder LC, Meyer MT, Griffiths H, Jonikas MC, Smith AM, & McCormick AJ.
Plant Biotechnology Journal (2015) 14:1302-1315
PMID: 26538195
Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome.
Jinkerson RE & Jonikas MC.
The Plant Journal (2015) 82: 393–412.
PMID: 25704665
Terashima M, Freeman ES, Jinkerson RE, & Jonikas MC.
The Plant Journal (2015) 81: 147-59.
PMCID: PMC4280329
Ion antiport accelerates photosynthetic acclimation in fluctuating light environments.
Armbruster U, Carrillo LR, Venema K, Pavlovic L, Schmidtmann E, Kornfeld A, Jahns P, Berry JA, Kramer DM, & Jonikas MC.
Nature Communications (2014) 5: 5439.
PMCID: PMC4243252
Yang W, Catalanotti C, D'Adamo S, Wittkopp TM, Ingram-Smith CJ, Mackinder L, Miller TE, Heuberger AL, Peers G, Smith KS, Jonikas MC, Grossman AR, & Posewitz MC.
The Plant Cell (2014) 26: 4499-4518.
PMCID: PMC4277214
Li X, Umen JG, & Jonikas MC.
Proceedings of the National Academy of Sciences U S A (2014) 111: 15610-15611.
PMCID: PMC4226088
Actin is required for IFT regulation in Chlamydomonas reinhardtii.
Avasthi P, Onishi M, Karpiak J, Yamamoto R, Mackinder L, Jonikas MC, Sale WS, Shoichet B, Pringle JR, & Marshall WF.
Current Biology. (2014) 24: 2025-2032.
PMCID: PMC4160380
*Zhang R, *Patena W, Armbruster U, Gang SS, Blum SR, & Jonikas MC.
*Equal contribution
The Plant Cell (2014) 26: 1398-1409.
PMCID: PMC4036561
Martin's previous work:
*Shurtleff MJ, *Itzhak DN, Hussmann JA, Schirle Oakdale NT, Costa EA, Jonikas M, Weibezahn J, Popova KD, Jan CH, Sinitcyn P, Vembar SS, Hernandez H, Cox J, Burlingame AL, Brodsky JL, Frost A, Borner GH, & Weissman JS.
*Equal contribution
eLife (2018) doi: 10.7554/eLife.37018.
PMID: 29809151
Automated identification of pathways from quantitative genetic interaction data.
Battle A, Jonikas MC, Walter P, Weissman JS, & Koller D.
Molecular Systems Biology (2010) 6: 379.
PMCID: PMC2913392
J domain co-chaperone specificity defines the role of BiP during protein translocation.
Vembar SS, Jonikas MC, Hendershot LM, Weissman JS, & Brodsky JL.
Journal of Biological Chemistry (2010) 285: 22484-22494.
PMCID: PMC2903355
Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum.
Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, & Schuldiner M.
Science (2009) 323: 1693-1697.
PMCID: PMC2877488
Identification of yeast proteins necessary for cell-surface function of a potassium channel.
Haass FA, Jonikas M, Walter P, Weissman JS, Jan YN, Jan LY, & Schuldiner M.
Proceedings of the National Academy of Sciences U S A (2007) 104: 18079-18084.
PMCID: PMC2084299